Parkinson’s disease (PD) is the second most common neurodegenerative disorder, characterized by the degeneration of dopamine-releasing regions in the brain. The loss of dopamine leads to a chronic and progressive decrease in movement, coordination, and cognition. While there is no cure, an early diagnosis promotes treatment efficacy and prolongs a higher quality of life.
Traditional research methods
Traditional stereological methods in PD research are time-consuming and involve tedious manual work, sometimes leading to error-prone analysis. One of the most time-consuming aspects is cell counting, specifically counting neurons positive for tyrosine hydroxylase (TH), a marker for dopaminergic neurons. This is also difficult with traditional, manual methods and involves inter- and intra-observer bias.
Artificial Intelligence (AI)- assisted image analysis increases project speed and scalability, saving researchers valuable time to focus their efforts on more complex and higher-level challenges. Large data management and analysis consistency eliminate variation from random sampling, leading to improved statistical accuracy and reproducible results for future PD research.
Christina Caira, PhD Student at Duke University in the Pharmacology and Cancer Biology Program, discusses the building of an AI model for TH quantification using Aiforia® Create in her PD research. Read her interview below.
Describe the project or research you are working on
"Broadly, our lab uses preclinical models (mouse and rat) to identify key mechanisms underlying neurodegenerative diseases like Parkinson’s disease and develop therapeutics that block disease progression. The pathological hallmarks of Parkinson’s disease are the accumulation of alpha-synuclein-containing inclusions called Lewy bodies and the loss of dopaminergic neurons.
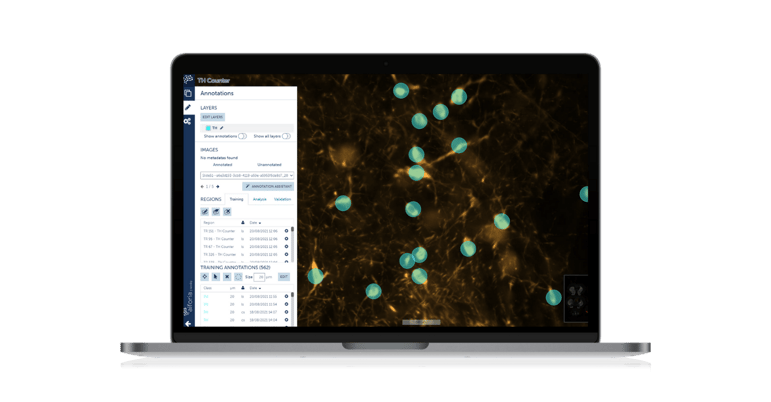
Using immunohistochemistry, we use antibodies to mark certain antigens (proteins) in cells of brain tissue to essentially quantify disease progression in our models. In these disease models, we can manipulate brain circuits hypothesized to be involved in pathogenesis using pharmacological drugs and compare them to appropriate controls. This gives us both mechanistic and potential therapeutic insight. The TH counter we built quantifies cells that are positive for tyrosine hydroxylase, which is a marker for dopaminergic neurons. The greater the reduction in TH+ cells, the greater the degeneration."
"I expected that using AI would help reduce some of the inherent biases and errors of manual counting. I also expected that it would reduce overall analysis time and offload the most tedious aspect of analysis, which is manual counting. Finally, I expected that it would help us create a more efficient workflow from experiment to analysis to communication of these results."
How has it been getting started with Aiforia?
"We have had an amazing experience with Aiforia thus far. Before Aiforia, I spent days on online help forums trying to resolve issues with file uploads, data storage, accurate analyses, etc. I often felt frustrated with the digital analysis interfaces and would end up manually counting tissue anyway because I didn’t trust the results. The support team at Aiforia has met with me frequently to help with questions and create protocols that cater to our specific file types. Their online resources are comprehensive and easy to understand; they really make AI accessible to people of all educational backgrounds (i.e. non-computer science backgrounds)."
How is the AI model performing compared to using traditional methods?
"The AI model is incredibly fast. Of course, there is a (reasonable) time and effort investment on the front end, but once the model is built and validated, analyses take only a few hours (where it used to take us a few weeks or even months). This speed has enabled us to add additional markers to our panels, which allows us to get the most from every animal used in our experiments. Additionally, the model is extremely accurate; we have validated it against human counting, and the model boasts an F1-score percentage over ninety-nine percent."
What are your next steps after this publication; do you plan to use AI for further research or other projects?
"This TH counter will be incorporated into pretty much every future project in our lab; we use tyrosine hydroxylase as an assessment of neurodegeneration in every model. Additionally, we are making AI models for every immunohistochemical marker used in our lab, such as alpha-synuclein, 6-diamidino-2-phenylindole (DAPI), neuronal nuclear protein (NeuN), ionized calcium-binding adaptor molecule 1 (Iba1), etc. We have also begun work on models for calculating fluorescent intensity of specific markers in brain regions of interest."
Read more neuroscience case studies: